Chapter 4 Missing Value
All the missing values come from one column: HOSP_COUNT_7DAY_AVG. The total amount of missing value is 3935 for this dataset.
colSums(is.na(data_by_day)) %>%
sort(decreasing = TRUE)## HOSP_COUNT_7DAY_AVG date_of_interest
## 3935 0
## areas CASE_COUNT
## 0 0
## PROBABLE_CASE_COUNT HOSPITALIZED_COUNT
## 0 0
## DEATH_COUNT PROBABLE_DEATH_COUNT
## 0 0
## CASE_COUNT_7DAY_AVG ALL_CASE_COUNT_7DAY_AVG
## 0 0
## DEATH_COUNT_7DAY_AVG ALL_DEATH_COUNT_7DAY_AVG
## 0 0
## PROBABLE_CASE_COUNT_7DAY_AVG HOSPITALIZED_COUNT_7DAY_AVG
## 0 0Using groupby areas to see the distribution of missing value:
data_by_day %>%
select(HOSP_COUNT_7DAY_AVG, areas) %>%
group_by(areas) %>% miss_var_summary()## # A tibble: 5 × 4
## # Groups: areas [5]
## areas variable n_miss pct_miss
## <chr> <chr> <int> <dbl>
## 1 BX HOSP_COUNT_7DAY_AVG 787 100
## 2 BK HOSP_COUNT_7DAY_AVG 787 100
## 3 MN HOSP_COUNT_7DAY_AVG 787 100
## 4 QN HOSP_COUNT_7DAY_AVG 787 100
## 5 SI HOSP_COUNT_7DAY_AVG 787 100gg_miss_var(data_by_day,
facet = areas)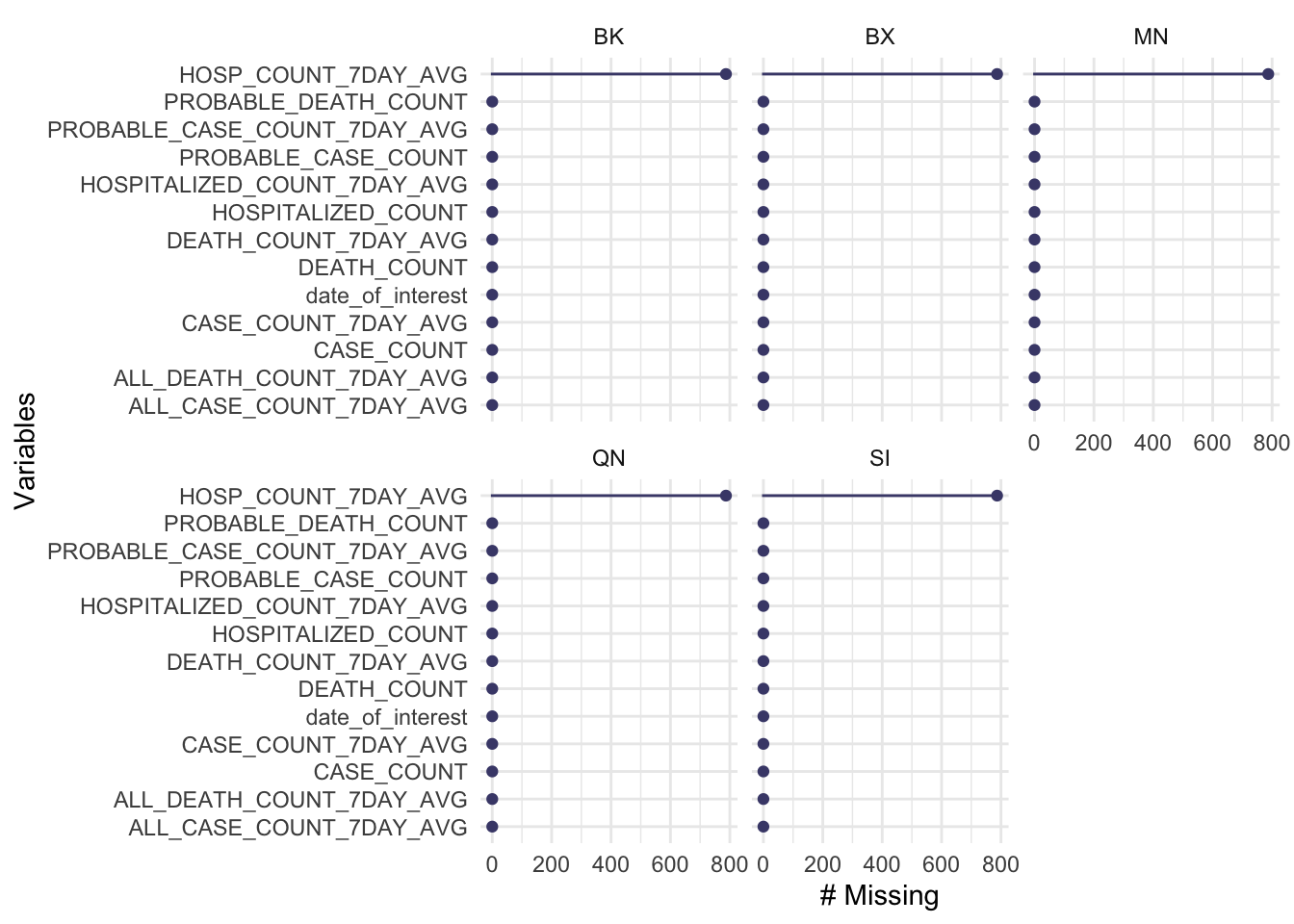
The missing values are equally distributed among BX, BK, MN, QN, SI counties, which more likely to be happen on the same date.
missing_range <-data_by_day %>%
select(HOSP_COUNT_7DAY_AVG, date_of_interest) %>%
group_by(date_of_interest) %>% miss_var_summary()head(missing_range)## # A tibble: 6 × 4
## # Groups: date_of_interest [6]
## date_of_interest variable n_miss pct_miss
## <chr> <chr> <int> <dbl>
## 1 02/29/2020 HOSP_COUNT_7DAY_AVG 5 100
## 2 03/01/2020 HOSP_COUNT_7DAY_AVG 5 100
## 3 03/02/2020 HOSP_COUNT_7DAY_AVG 5 100
## 4 03/03/2020 HOSP_COUNT_7DAY_AVG 5 100
## 5 03/04/2020 HOSP_COUNT_7DAY_AVG 5 100
## 6 03/05/2020 HOSP_COUNT_7DAY_AVG 5 100tail(missing_range)## # A tibble: 6 × 4
## # Groups: date_of_interest [6]
## date_of_interest variable n_miss pct_miss
## <chr> <chr> <int> <dbl>
## 1 04/20/2022 HOSP_COUNT_7DAY_AVG 5 100
## 2 04/21/2022 HOSP_COUNT_7DAY_AVG 5 100
## 3 04/22/2022 HOSP_COUNT_7DAY_AVG 5 100
## 4 04/23/2022 HOSP_COUNT_7DAY_AVG 5 100
## 5 04/24/2022 HOSP_COUNT_7DAY_AVG 5 100
## 6 04/25/2022 HOSP_COUNT_7DAY_AVG 5 100The missing value occurs from 02/29/2020 to 04/25/2022. Between this time frame, varaible HOSP_COUNT_7DAY_AVG records are missing over the 5 counties.
To deal with those missing value, as we are not quite interested in variable HOSP_COUNT_7DAY_AVG, we will not be bothered by the existence of missing values and will keep all the records in the dataset to continue further analysis.
Consider about the correlation between the variables in the dataset:
library(corrplot)
library(RColorBrewer)
source("http://www.sthda.com/upload/rquery_cormat.r")
mydata <- data_by_day[, c(3:8)]
require("corrplot")
rquery.cormat(mydata)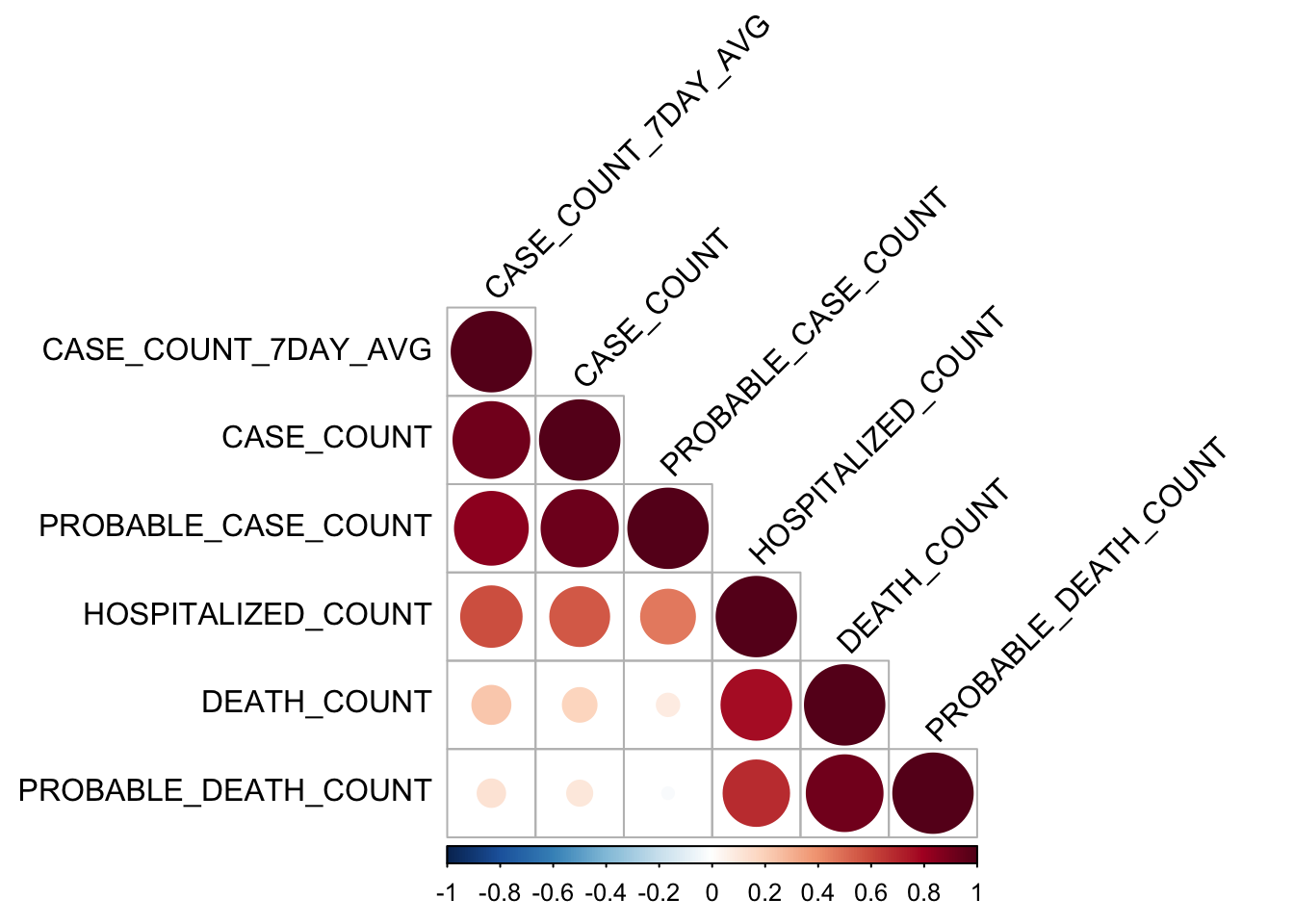
## $r
## CASE_COUNT_7DAY_AVG CASE_COUNT PROBABLE_CASE_COUNT
## CASE_COUNT_7DAY_AVG 1
## CASE_COUNT 0.91 1
## PROBABLE_CASE_COUNT 0.84 0.92 1
## HOSPITALIZED_COUNT 0.58 0.55 0.46
## DEATH_COUNT 0.23 0.18 0.08
## PROBABLE_DEATH_COUNT 0.12 0.1 -0.022
## HOSPITALIZED_COUNT DEATH_COUNT PROBABLE_DEATH_COUNT
## CASE_COUNT_7DAY_AVG
## CASE_COUNT
## PROBABLE_CASE_COUNT
## HOSPITALIZED_COUNT 1
## DEATH_COUNT 0.77 1
## PROBABLE_DEATH_COUNT 0.68 0.91 1
##
## $p
## CASE_COUNT_7DAY_AVG CASE_COUNT PROBABLE_CASE_COUNT
## CASE_COUNT_7DAY_AVG 0
## CASE_COUNT 0 0
## PROBABLE_CASE_COUNT 0 0 0
## HOSPITALIZED_COUNT 0 1.9e-308 1.1e-208
## DEATH_COUNT 1.5e-49 1.4e-31 4.4e-07
## PROBABLE_DEATH_COUNT 8.8e-15 2.3e-10 0.17
## HOSPITALIZED_COUNT DEATH_COUNT PROBABLE_DEATH_COUNT
## CASE_COUNT_7DAY_AVG
## CASE_COUNT
## PROBABLE_CASE_COUNT
## HOSPITALIZED_COUNT 0
## DEATH_COUNT 0 0
## PROBABLE_DEATH_COUNT 0 0 0
##
## $sym
## CASE_COUNT_7DAY_AVG CASE_COUNT PROBABLE_CASE_COUNT
## CASE_COUNT_7DAY_AVG 1
## CASE_COUNT * 1
## PROBABLE_CASE_COUNT + * 1
## HOSPITALIZED_COUNT . . .
## DEATH_COUNT
## PROBABLE_DEATH_COUNT
## HOSPITALIZED_COUNT DEATH_COUNT PROBABLE_DEATH_COUNT
## CASE_COUNT_7DAY_AVG
## CASE_COUNT
## PROBABLE_CASE_COUNT
## HOSPITALIZED_COUNT 1
## DEATH_COUNT , 1
## PROBABLE_DEATH_COUNT , * 1
## attr(,"legend")
## [1] 0 ' ' 0.3 '.' 0.6 ',' 0.8 '+' 0.9 '*' 0.95 'B' 1It is quite obvious to see that all cases are positive correlated and the correlation is quite strong for the count of cases and their 7 day count cases under the same category.